
Thousands of protein switches enable plants to make the transition from dark to light
We already learn in school that plants carry out photosynthesis in their chloroplasts when exposed to light. In the dark, this process comes to a halt. Instead, green plant tissue – for example the leaves – then activate cellular respiration, during which they consume carbohydrates and oxygen in the mitochondria, similarly to what humans are constantly doing. These two processes – photosynthesis and cellular respiration – take place separately in plant cells, both spatially and temporally. The fundamental switch which occurs in their metabolism is a tour de force which plants perform almost unnoticed – and in a very short time span – in the daily transition from light to dark.
What actually happens in the plant at the molecular level is something that has kept researchers busy for decades now. However, most studies so far focused on the regulation of individual proteins. Now a research team headed by Prof. Iris Finkemeier from the Institute of Biology and the Biotechnology of Plants at the University of Münster has published a study which analyses the central reprogramming at the protein level in an unprecedented sharpness of detail: the researchers analysed leaf tissue from plants during the transition from darkness to light and were able to confirm that the photosynthesis process was fully active within just 15 minutes after the onset of light. In doing so, the plant does not produce its entire stock of proteins anew, but rather undertakes targeted biochemical changes on the proteins which already exist – so-called post-translational modifications (PTMs for short). These PTMs can then, in turn, control the activity of enzymes and ultimately lead to a rapid, fundamental switchover in the cell’s function. The results of the study have now been published in “The Plant Journal”.
Background and methods used
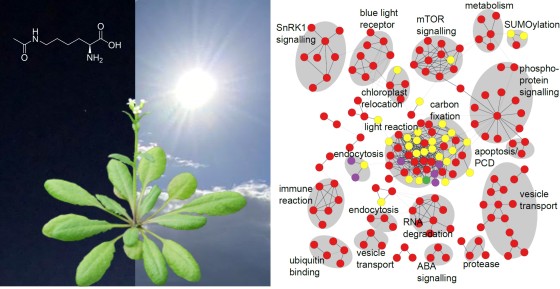
In addition to applying physiological methods to measure the level of photosynthesis and the metabolite composition, the team used high-resolution protein mass spectrometry in order to be able to observe several thousand of proteins simultaneously. Using mass spectrometry, scientists determine the exact mass of complex molecules. If the molecule has a chemical modification, it has a different mass from a molecule without such a modification. “We can determine, to an accuracy of one ppm, the exact mass of more than ten thousand peptide molecules in a sample,” says Dr. Jürgen Eirich, a co-author of the study. “This corresponds to a deviation of just one milligram per kilogram. We can compare this to weighing all the residents of a small town to an accuracy of a tenth of a gram.” The first changes could already be seen after five minutes’ exposure to light – although at this point most of the changes to the proteins were still taking place in the chloroplasts, i.e. where the start of the photosynthesis process leads primarily to changes in the cell’s function. These changes are not, however, restricted to the chloroplast. After 30 minutes, the entire cell is affected by a variety of changes to its proteins.
The results of the study are freely available for public use. They offer biologists and biotechnologists, as well as plant breeders, a rich basis for modifying plants’ metabolism at decisive points or for switching metabolic pathways on or off in order to improve the growth, yields or resilience of plants.
Other researchers from the University of Bonn and the Max Planck Institute of Molecular Plant Physiology in Golm were involved in the study. The work received financial support from the German Research Foundation.
Original publication
Giese, J. et al (2023). The interplay of posttranslational protein modifications in Arabidopsis leaves during photosynthesis induction. The Plant Journal. DOI: 10.1111/tpj.16406
