RESEARCH - Andrea Rentmeister
Biological Chemistry and Biomolecular Label Chemistry
ERC Grant RNActivate

Light is an excellent external regulatory element that can be applied to cells and organisms with high spatio-temporal precision and without interfering with cellular processes. Optochemical biology exploits small photo-responsive chemical groups to cage and activate or to switch biomolecular functions in response to light of a defined wavelength. Caged antisense agents have enabled down-regulation of gene expression with spatio-temporal control at the messenger-RNA (mRNA) level in vivo, however approaches for triggering translation of exogenous mRNA lack efficient turn-on effects. To explore the effects of conditional and transient ectopic gene expression in a developing organism it is vital to fully abrogate and restore translational efficiency.
The goal of this project is to bring eukaryotic mRNA under the control of light to trigger efficient ectopic translation with spatio-temporal resolution in cells and in vivo. In addition to labeling and tracking subpopulations of cells, we will use our technology to control and to manipulate cell fate by locally producing proteins responsible for cell death, genome engineering, and cell migration. We will use cultured cells and one-cell stage zebrafish embryos that can be easily injected with mRNA to study the function of ectopic gene expression in early development. Our approach will overcome current limitations of photo-inducible mRNA translation and enable us to manipulate a developing organism at the molecular level.
Background
The 5' cap as a regulator of translation
Eukaryotic mRNAs have two major hallmarks, namely the 5' cap and the poly(A) tail. The 5' cap consists of N7-methylguanosine that is connected to the 5' end of an mRNA via an unusual 5'-5' triphosphate bridge (Fig. 1A). The cap is canonically made co-transcriptionally in the nucleus by the consecutive action of three enzymes, namely a triphosphatase, a guanylyltransferase, and an N7 methyltransferase (MTase).[1]
The 5' cap serves as an interaction platform for several proteins and is important for mRNA splicing, export, protection from exoribonucleases, and translation. In the cytoplasm the 5' cap is bound by the eukaryotic translation initiation factor (eIF4E) and this direct binding is thought to be the rate-limiting step in translation initiation (Fig. 1B). The importance of the 5' cap for translation and its potential for transcript stabilization have prompted fundamental efforts to engineer and synthesize modified cap analogs with increased stability and translational efficiency for applications as mRNA therapeutics, e.g. mRNA-based cancer immunotherapy.[2]
The crystal structure of eIF4E in complex with m7GTP reveals that tryptophan stacking of W56 and W102 with the guanosine and a salt bridge between N2 and E103 are essential for recognition (Fig. 3B). The protein eIF4E can discriminate between the canonical m7Gppp-mRNA and Gppp-mRNA lacking the N7 methyl group by a remarkable factor of >100 because the delocalized positive charge arising from the N7 methyl group further strengthens the interactions between the π-electrons of the stacked aromatic rings. The methyl group of m7G is additionally stabilized by van der Waals interactions with W166.[1]

Methodology
The 5' cap/eIF4E interaction is highly sensitive to modifications at position N7.[4] To produce mRNAs with different modifications at the N7 position of the 5' cap, we developed an efficient enzymatic approach based on the highly promiscuous MTase Ecm1 (Fig. 1A). We successfully synthesized AdoMet analogs and used them to modify the N7 position of the cap with sterically demanding groups, including benzylic moieties, such as vinylbenzyl, azidobenzyl, and diazirine in quantitative conversions.[3a] Even benzophenone was transferred, albeit with a lower yield (60 %).[3a] The only prerequisite for transfer is an unsaturated C-atom in β-position, required for transition state stabilization.[5] We incorporated the modified 5' caps into reporter-mRNAs and showed that even small N7-modifications like the allyl group efficiently blocked translation of reporter-mRNAs in vitro and in mammalian cells (Fig. 1C, D).[3b] Furthermore, we achieved intracellular labeling of these modified mRNAs in living cells.[3b]
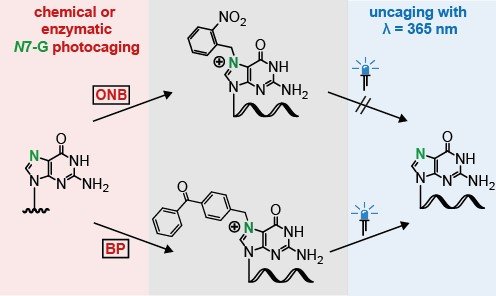
We have also developed methodology for the enzymatic photocaging of DNA as well as the 5' cap.[6] Interestingly, we found that typical photocaging groups like the ortho-nitrobenzyl (ONB) group are not suitable for the N7 position of guanosine. Here, we reported the benzophenone groups to be photocleavable (Figure 2).
Experimental Approach
- Chemo-enzymatic synthesis of mRNAs with 5' cap
- Translation studies in vitro and in cells
- Injection into zebrafish embryos (in collaboration with Prof. Erez Raz and Prof. Stefan Schulte-Merker, both WWU)
- Confocal laser scanning microscopy
People
- Andrea Rentmeister (PI)
- Petr Špaček (Postdoc)
- Nils Klöcker (PhD student)
- Melissa van Dülmen (PhD student)
- Florian Weißenböck (Master student)
- Oleksii Zozulia
- Amarnath Bollu
- Anne-Marie Lawrence-Dörner (TA)
References
[1] I. Topisirovic, Y. V. Svitkin, N. Sonenberg and A. J. Shatkin, Wiley Interdisciplinary Reviews-RNA 2011, 2, 277-298.
[2] a) J. Kowalska, A. Wypijewska del Nogal, Z. M. Darzynkiewicz, J. Buck, C. Nicola, A. N. Kuhn, M. Lukaszewicz, J. Zuberek, M. Strenkowska, M. Ziemniak, M. Maciejczyk, E. Bojarska, R. E. Rhoads, E. Darzynkiewicz, U. Sahin and J. Jemielity, Nucleic Acids Research 2014, 42, 10245-10264; b) U. Sahin, K. Kariko and O. Tureci, Nature Reviews Drug Discovery 2014, 13, 759-780.
[3] a) F. Muttach, F. Mäsing, A. Studer and A. Rentmeister, Chemistry - A European Journal 2017, accepted, doi:10.1002/chem.201605663; b) J. M. Holstein, L. Anhäuser and A. Rentmeister, Angewandte Chemie International Edition 2016, 55, 10899-10903.
[4] a) E. Darzynkiewicz, J. Stepinski, I. Ekiel, C. Goyer, N. Sonenberg, A. Temeriusz, Y. Jin, T. Sijuwade, D. Haber and S. M. Tahara, Biochemistry 1989, 28, 4771-4778; b) A. Niedzwiecka, J. Marcotrigiano, J. Stepinski, M. Jankowska-Anyszka, A. Wyslouch-Cieszynska, M. Dadlez, A. C. Gingras, P. Mak, E. Darzynkiewicz, N. Sonenberg, S. K. Burley and R. Stolarski, Journal of Molecular Biology 2002, 319, 615-635.
[5] C. Dalhoff, G. Lukinavicius, S. Klimasauskas and E. Weinhold, Nature Chemical Biology 2006, 2, 31-32.
[6] a) L. Anhäuser, N. Klöcker, F. Muttach, F. Masing, P. Spacek, A. Studer and A. Rentmeister, Angew Chem Int Ed Engl 2019, 10.1002/anie.201914573; b) L. Anhäuser, F. Muttach and A. Rentmeister, Chemical Communications 2018, 54, 449-451.
Publications from ERC grant
Anhäuser, L., Klöcker, N., Muttach, F., Mäsing, F., Spacek, P., Studer, A., Rentmeister, A. (2020)
A benzophenone-based photocaging strategy for the n7 position of guanosine.
Angew. Chem. Int. Ed., 59(8):3161-3165. Open Access Link
Cornelissen, N.V., Michailidou, F., Muttach, F., Rau, K., Rentmeister, A. (2020)
Nucleoside-modified adomet analogues for differential methyltransferase targeting.
ChemCommun. 56(14): 2115-2118. Open Access Link
Klöcker, N., F. P. Weissenboeck and A. Rentmeister (2020)
Covalent labeling of nucleic acids.
Chemical Society Reviews. Open Access Link
Michailidou, F., N. Klöcker, N. Cornelissen, R. K. Singh, A. Peters, A. Ovcharenko, D. Kümmel and A. Rentmeister (2021)
Engineered SAM synthetases for enzymatic generation of AdoMet analogs with photocaging groups and reversible DNA modification in cascade reactions.
Angew. Chem. Int. Ed., 60:480-485. Open Access Link
Ovcharenko, A., Weissenboeck, F.P., and Rentmeister, A. (2021)
Tag-Free Internal RNA Labeling and Photocaging Based on mRNA Methyltransferases.
Angew. Chem. Int. Ed., 60(8):4098-4103. Open Access Link
Westerich, K.J., Chandrasekaran, K.S., Gross-Thebing, T., Kück, N., Raz, E., Rentmeister, A. (2020)
Bioorthogonal mRNA labeling at the poly(A) tail for imaging localization and dynamics in live zebrafish embryos.
Chemical Science 11(11):3089-3095. Open Access Link