RESEARCH - Andrea Rentmeister
CRC1459: Intelligent matter: From responsive to adaptive nanosystems
https://www.uni-muenster.de/SFB1459/
Our project: A semi-synthetic nanosystem for programmable control of output based on rational design and directed evolution
In this project, we aim to develop a system that senses different small molecules and actuates the formation of an output signal. The semi-synthetic nanosystem consists of a reaction network in aqueous solution and explores different feedback mechanisms to move from a responsive to an adaptive system that self-regulates the formation of metabolites, RNA, or protein and leads to a detectable output signal (e.g. fluorescence, enzymatic activity or proteinaceous material). Permanent or light-removable modifications of DNA will be tested as a molecular memory indicating which fuels the system has previously been exposed to. The long-term goal of this project is to realize adaptive behavior and learning capability, reminiscent of artificial synapses, by integrating this sensing-actuation system into photonic circuits or confined spaces, such as vesicles and nanofabrication approaches, and to be able to distinguish experienced systems from naïve ones.
General background: Actuation by methylation
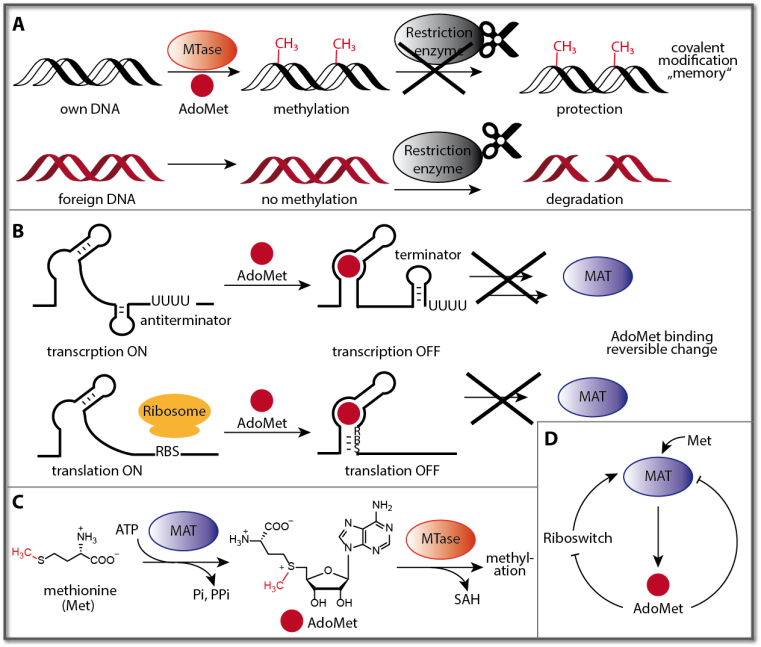
Methylation is a small but important natural modification regulating the activity of small biomolecules and all classes of biopolymers. Methylation of DNA and RNA is responsible for actuating multiple biological processes, with epigenetic silencing of genes by methylation of CpG sites in promoters as most impressive example. Bacteria use methylation of DNA as defense mechanism (Fig. 1A). They mark their own DNA using sequence-specific methyltransferases (MTases) to distinguish it from foreign invading phage DNA, which is cut by endogenous restriction enzymes.
The methyl group originates in most cases from S-adenosyl-L-methionine (SAM or AdoMet) – the second most abundant co-factor in nature – which is made from methionine and ATP by methionine adenosyltransferase (MAT), a ubiquitous enzyme (Fig. 1C). In nature, methionine or AdoMet thus serve as an input or fuel that is sensed and actuates a response or output, either by reversible changes (binding) or permanent methylation. The output can be differences in protein production or accessibility of genomic information in DNA. All processes involving methylation and demethylation as well as the intracellular level of AdoMet are tightly regulated, using feedback at different levels (Fig. 1D). Most MATs show strong product inhibition by AdoMet (feedback inhibition). In bacteria, expression of genes involved in AdoMet synthesis – including MAT – are regulated by SAM riboswitches on the level of transcription (SAM I riboswitch) and translation (SAM II – VI riboswitches) (Fig. 1B). This feedback mechanism attenuates the expression of genes involved in the AdoMet synthesis (e.g. MAT) in response to high AdoMet levels. Riboswitches are typically found in the 5′-untranslated region (5′-UTR) of the genes they are regulating. These RNAs consist of an aptamer domain (responsible for binding) and an expression domain (responsible for regulation). When the ligand – in this case AdoMet – binds to the aptamer domain, changes in the secondary structure occur that also affect the expression domain. In the case of transcriptional regulation, a terminator stem is formed, which causes transcription to stop (Fig. 1B). Similarly, in the case of translational regulation, the ribosome-binding site (RBS) is sequestered and the ribosome cannot bind any more, preventing translation (Fig. 1B). To date, six classes of SAM riboswitches are known (SAM I to SAM VI). This is the highest number of riboswitches known for a single metabolite, underscoring the importance of regulating gene expression in response to AdoMet levels.
Our work
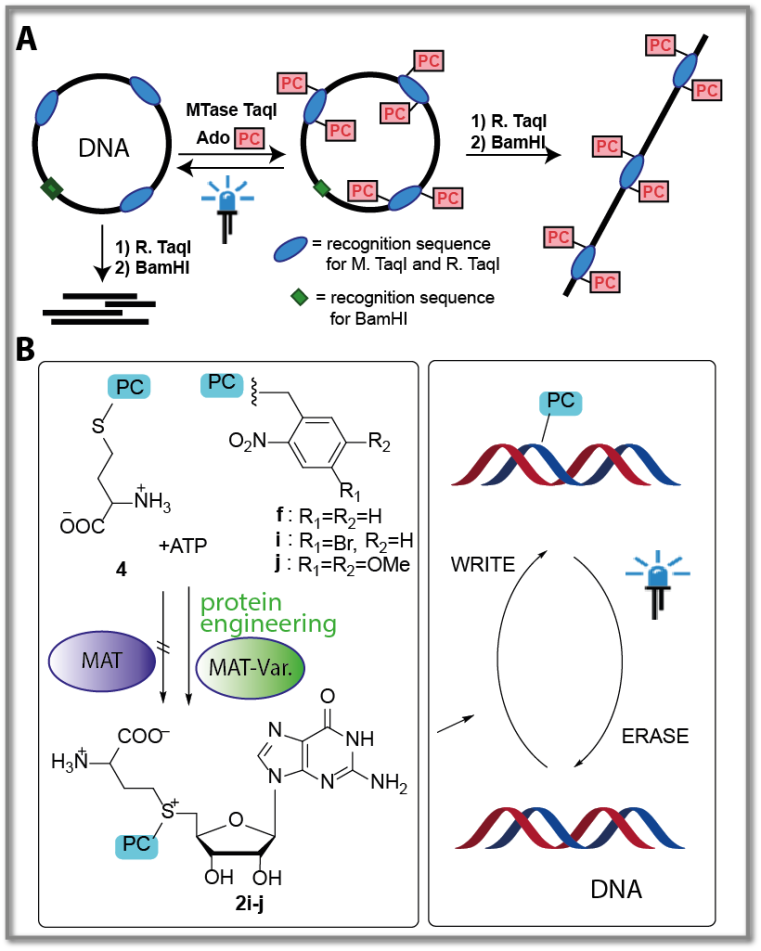
We recently established reversible modification of DNA by sequence-specific enzymatic transfer of photo-caging groups using the promiscuous MTase MTaqI [L. Anhauser, 2018] (Fig. 3A). The plasmid DNA was then treated by two restriction enzymes (RTaqI and BamHI) and degraded, if it was unmodified. When the plasmid was modified (either methylated or with photo-caging groups), the DNA was only linearized but not degraded (Fig. 3A). The photo-caging groups could then be removed by short irradiation with UV light, allowing subsequent degradation to occur. This is a first example of controlling the natural methylation/restriction system by non-natural substrates and light.
A limitation of AdoMet and AdoMet analogs is their limited stability in aqueous solution. To address this limitation, we established their enzymatic generation from methionine analogs and ATP using MAT. We set up an enzymatic cascade for in situ generation of AdoMet analogs with longer alkyl side chains and direct transfer to the 5’ cap (Fig. 2D) [F. Muttach, 2016]. However, benzylic residues – which are the basic building block of typical photo-caging groups, such as the ortho-nitrobenzyl group – were not accepted by WT MAT enzymes. To address this issue, we started to engineer a MAT enzyme based on structure-guided mutations. We are now able to convert also methionine analogs with a benzylic or a photo-caging group based on the ortho-nitrobenzyl (ONB) moiety (4f, i, j), which has not been reported to the best of our knowledge (Fig. 3B, Michailodou et al, Angew 2021). These variants provide the starting point in this project for further engineering of MAT to efficiently convert photo-caging groups.
Publications
Rickhoff, J., N. V. Cornelissen, T. Beuse, A. Rentmeister and B. Jan Ravoo (2021).
"Multiresponsive hydrogels and organogels based on photocaged cysteine."
Chemical Communications.
https://pubs.rsc.org/en/content/articlelanding/2021/cc/d1cc01363g#!divAbstract
Michailidou, F., N. Klöcker, N. V. Cornelissen, R. K. Singh, A. Peters, A. Ovcharenko, D. Kümmel and A. Rentmeister (2021).
"Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions."
Angewandte Chemie International Edition 60(1): 480-485.
Muttach F, Rentmeister A. (2016).
A Biocatalytic Cascade for Versatile One-Pot Modification of mRNA Starting from Methionine Analogues.
Angew Chem Int Ed Engl. Jan 26;55(5):1917-20. doi: 10.1002/anie.201507577. Epub 2015 Dec 22. PMID: 26694947.