RESEARCH - Andrea Rentmeister
Biological Chemistry and Biomolecular Label Chemistry
Non-natural amino acids for probing methylation sites in vitro and in cells
N6-Methyladenosine (m6A) is the most abundant internal modification in eukaryotic mRNA. It is introduced by METTL3-METTL14 and tunes mRNA metabolism, impacting cell differentiation and development. Precise transcriptome-wide assignment of m6A sites is of utmost importance. However, m6A does not interfere with Watson-Crick base pairing making polymerase-based detection challenging. We developed a chemical biology approach for the precise mapping of methyltransferase (MTase) target sites based on the introduction of a bioorthogonal propargyl group in vitro and in cells. We showed that propargyl can be introduced enzymatically by wild-type METTL3-METTL14. Reverse transcription terminated up to 65 % at m6A sites after bioconjugation and purification, hence enabling detection of METTL3-METTL14 target sites by next generation sequencing.
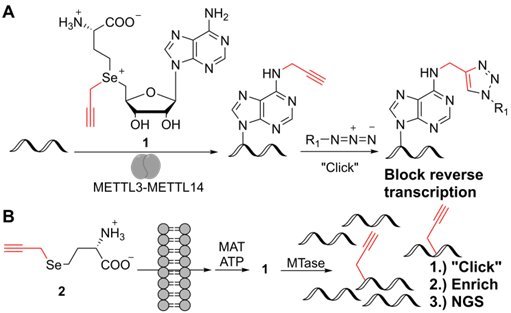
Importantly, we tested whether our strategy can be used to identify RNA MTase target sites in vivo. To this end, we established metabolic labeling of MTase target sites via biosynthesis of AdoMet analogs from methionine analogs and ATP by methionine adenosyltransferase (MAT) (Figure 1B). Specifically, we fed mammalian HeLa cells with propargyl-L-selenohomocysteine (2), then isolated total RNA and validated formation of 2'-O-propargyl-adenosine by LC-MS/MS. Next, we clicked the total RNA to a biotin-azide and generated NGS libraries. We mapped the reads to ribosomal RNA, which is known to contain different types of nucleotide methylations. We assigned peaks enriched in RNA from metabolic propargyl labeling as well as RNA from control cells and compared their position to known methylation sites in human rRNA.
In combination with bioconjugation and NGS, our approach presents a new avenue for direct and precise assignment of m6A sites by blocking reverse transcription independent of potential antibody biases. Compared to a recently described method, based on iodination of N6-allyladenosine, our approach has several advantages: (i) the CuAAC reaction is highly selective and enables RNA enrichment, (ii) the propargyl group is more efficiently introduced than the allyl group, because we use a selenium-based AdoMet analog and (iii) we could introduce non-natural modifications in vivo. For the first time, we show that a propargyl-group can be used for metabolic labeling of RNA MTase target sites by feeding cells with the respective amino acid precursor 2. In combination with our bioconjugation and NGS protocol, we confirmed known MTase target sites in rRNA. Adaptations for mRNA sequencing will likely enable detection of m6A sites in vivo. Finally, our strategy paves the way to identify target sites of different RNA MTases in a transcriptome-wide manner.
Selected references
Michailidou, F., N. Klöcker, N. Cornelissen, R. K. Singh, A. Peters, A. Ovcharenko, D. Kümmel and A. Rentmeister (2021)
Engineered SAM synthetases for enzymatic generation of AdoMet analogs with photocaging groups and reversible DNA modification in cascade reactions.
Angew. Chem. Int. Ed., 60(1):480-485. Open Access Link
Ovcharenko, A., Weissenboeck, F.P., and Rentmeister, A. (2021)
Tag-Free Internal RNA Labeling and Photocaging Based on mRNA Methyltransferases.
Angew. Chem. Int. Ed., 60(8):4098-4103. Open Access Link
Hartstock, K., Nilges, B. S., Ovcharenko, A., Cornelissen, N. V., Püllen, N., Lawrence-Dörner, A., Leidel, S. A., and Rentmeister, A. (2018)
Enzymatic or in vivo installation of propargyl groups in combination with click chemistry for the enhancement and detection of methyltransferase target sites in RNA
Angew. Chem. Int. Ed. 2018, 57:6342-6346. doi:10.1002/anie.201803995
Muttach, F., and Rentmeister A. (2016)
A biocatalytic cascade for versatile one-pot modification of mRNA starting from methionine analogues
Angew. Chem. Int. Ed., 55:1917-1920.