RESEARCH - A. Rentmeister
Biological Chemistry and Biomolecular Label Chemistry
Chemo-enzymatic modification of mRNA
Eukaryotic messenger RNA (mRNA) has emerged as a valuable alternative to DNA-based vectors for exogenous protein expression in eukaryotic cells. Benefits include the almost direct start of translation and typically higher transfection efficiencies because delivery to the cytoplasm is sufficient. Although low stability of mRNA compared to DNA can be an issue, the fact that mRNA does not permanently alter the genomic setting of the cell is now considered advantageous for therapeutic applications. Consequently, approaches to tune the translation efficiency and stability of specific transcripts are attracting more and more interest.
One crucial element for translation initiation of mRNAs is the interaction of the 5'-cap with the eukaryotic translation initiation factor eIF4E and alterations of the 5'-cap can directly impact translation. In addition to their general importance for translation, mRNAs can be asymmetrically distributed in eukaryotic cells and in several cases even locally translated. However, the molecular details of mechanisms such as active transport leading to subcellular localization are poorly understood. To investigate these dynamic processes, methods for labeling mRNAs in living cells are required. Ideally, the mRNA of interest should be altered as little as possible.
We developed a chemo-enzymatic approach to label in vitro produced and capped RNAs at the N2 position of the 5'-cap using a variant of the trimethylguanosine synthase from Giardia lamblia (GlaTgs2-V34A). However, the activity of this methyltransferase requires prior methylation of the N7 atom and even engineered variants showed compromised activity on larger analogs of the cosubstrate S-adenosyl-L-methionine (AdoMet) limiting the labeling yield to ~30%. Searching for a more straightforward and efficient approach, we noted Ecm1 - a cap (guanine N7) methyltransferase from the microsporidian parasite Encephalitozoon cuniculi - whose active site is located in a cleft rather than a binding pocket. We found that Ecm1 exhibits pronounced substrate promiscuity enabling fast and efficient conversion of a range of AdoMet analogs and thus makes Ecm1 an ideal enzyme for producing modified 5'-caps.
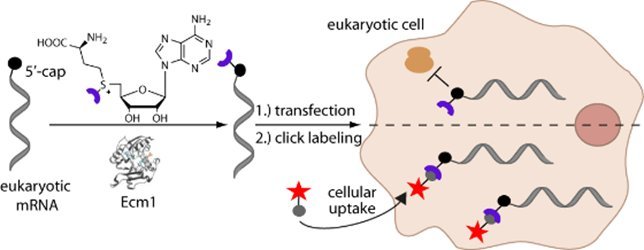
Methylation at position N7 is crucial for efficient translation and the N7 methylated cap is specifically and tightly bound by the eukaryotic translation initiation factor eIF4E. In addition to tuning translation, some of the non-natural cap modifications can be used for subsequent bioorthogonal reactions, potentially even for mRNA labeling in living cells. Intracellular labeling of a designated target mRNA that does not contain alterations of its nucleotide sequence, would give access to directly studying subcellular localization and trafficking of this mRNA at a given timepoint.
Therefore, we developed an enzymatic approach that provides access to tuning the translation efficiency and to intracellular labeling of a specific mRNA in living cells. The efficient transfer enables for the first time easy and straightforward enzymatic production of long translatable cap-modified mRNAs as exemplified for the eGFP- and the luciferase mRNAs, which are ~1000-2000 nt long. Tuning translation by cap modification allows to study functions of a target mRNA beyond serving as a template for translation. In combination with bioorthogonal click chemistry, we provide a tool for intracellular labeling of a target mRNA which will be valuable to study subcellular localization during dynamic processes such as outgrowth of polarized cells or development.
Selected references:
van Dülmen, M., N. Muthmann and A. Rentmeister (2021)
Chemo-enzymatic modification of the 5' cap maintains translation and increases immunogenic properties of mRNA.
Angewandte Chemie International Edition, in press, Open Access Link
Reichert, D., Mootz, H.D., and Rentmeister, A. (2021)
Light-control of cap methylation and mRNA translation via genetic code expansion of Ecm1.
Chem. Sci., 12(12):4383-4388. Open Access Link
Muttach, F., Muthmann, N., Reichert, D., Anhäuser, L., and Rentmeister, A. (2017)
A benzylic linker promotes methyltransferase catalyzed norbornene transfer for rapid bioorthogonal tetrazine ligation
Chemical Science, 8(12):7947-7953
Holstein, J.M., Anhäuser, L., and Rentmeister, A. (2016)
Modifying the 5'-cap for click reactions of eukaryotic mRNA and to tune translation efficiency in living cells
Angew Chem Int Ed Engl, 55:10899-903
Holstein, J.M., Stummer, D., and Rentmeister, A. (2015)
Enzymatic modification of 5'-capped RNA with a 4-vinylbenzyl group provides a platform for photoclick and inverse electron-demand Diels-Alder reaction
Chemical Science, 6(2):1362-1369
Schulz, D., Holstein, J.M., and Rentmeister, A. (2013)
A chemo-enzymatic approach for site-specific modification of the RNA cap
Angewandte Chemie International Edition, 52:7874-7878