2021
We are recruiting
Great to hear the positive feedback from DFG that the second phase of NC³ initiative is funded. It was a great experience during the last two years in the NC³ and we look forward to a fun and productive 4-year in NC³.
We are recruiting 1 PhD and 2 postdocs, working on single-cell genomics and experimental evolution. More information can be found here.
Join the group
PhD position (evolutionary genetics)
Postdoc position 1 (Evolutionary single cell genomics) - download call
Postdoc position 2 (Evolutionary ecology - download call
Move to Mainz
After nearly four years wonderful time at WWU Münster, we will move to JGU Mainz in April 2022. We will recruit new PhD students and postdocs soon.
A new paper!
Lanlan’s paper on the pleiotropic singling between flower development and herbivory-induced responses is accepted by FPS. Congratulations to Lanlan and other co-authors.
The paper can be accessed here
A new review paper!
A review paper from the duckweed community is now published in The Plant Cell. Here, we highlighted the great potential of using the duckweed as a model system to answer excellent questions in evolutionary ecology.
The paper can be accessed here

© Antonino Malacrinò Antonino Malacrinò joined the Xu lab

© Pablo Duchén Pablo Duchén joined the Xu lab
Congratulations to Thomas Schacker on his bachelor thesis
Thomas did a wonderful job for his BSc thesis. Congratulations and good luck in your further career, Thomas!
Congratulations to Samuel Wink on his bachelor thesis
Samuel successfully finished his BSc thesis in our group. He will further on stay with us as a student helper.
Congratulations, Samuel!
© Li et al. A paper published in Science

© Pascal Poweleit Pascal Poweleit joined group
2020
Great news!
Great news from the editor of Science. Excellent team work! Looking forward to seeing the paper in print soon.
A new project!
A new project is now funded by DFG. We will study the evolution of Colorado potato beetle and its insecticide resistance. A junior group leader position will be available soon.
Congratulations to our bachelors
Despite this difficult time, Fabio, Milena and Maximillan did wonderful job for their bachelor thesis. Congratulations to all of you!
New positions in plant evolution study funded by DFG
A new project is funded by DFG to study how microbes and herbivores together shape plant evolution. New postdoc and PhD positions will soon be available. If you are interested in joining the team, please contact us.

© Marie Sárazová Snails eat duckweeds, we eat cake

© Xu et al. 2020 Coevolution of floral signals and indirect defenses in plants

© Xu Lab Evolution of an adaptive and novel floral volatile
2019

© Xu lab Call for application: a field excursion in Southern Italy in 2020.
A Paper is accepted

© xu lab Xu group is climbing

© Althia Edison Alitha Edison joined the group
A new project in the group is funded by DFG
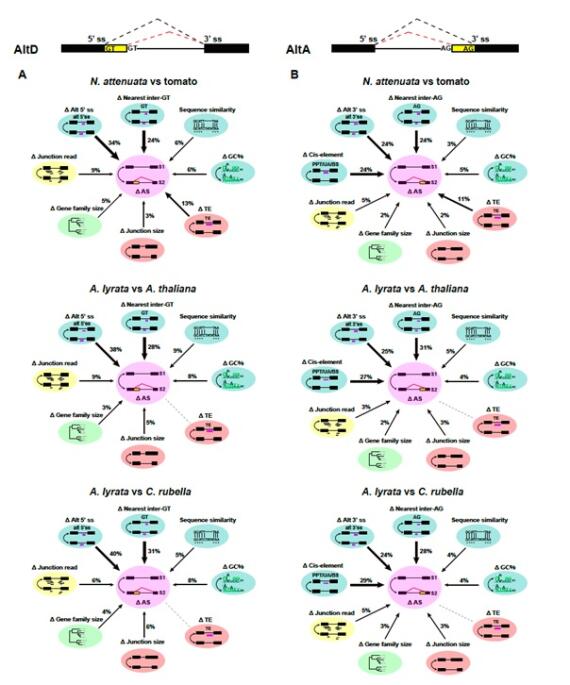
© Shuqing Xu New publication on alternative splicing evolution in plants

© Shuqing Xu Our first team building event
© Marie Sarazova & Yangzi Wang Two new members join the group
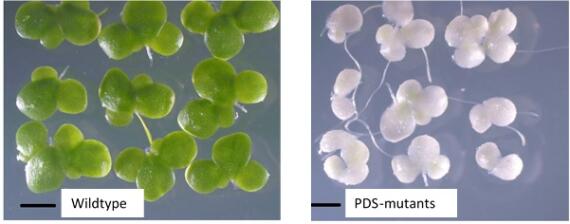
© Yubin Ma Efficient genetic transformation and CRISPR/Cas9‐mediated genome editing in Lemna aequinoctialis

© Klaus J. Appenroth Duckweed: The low-down on a tiny plant
2018

© Lanlan Ke Lanlan Ke joins XuLab!
Dr. Martin Schäfer has joined the group.

© XuLab Xu lab started officially at the Institute of Evolution and Biodiversity, University of Münster.


