1. Adaptation-in-action to herbicides.
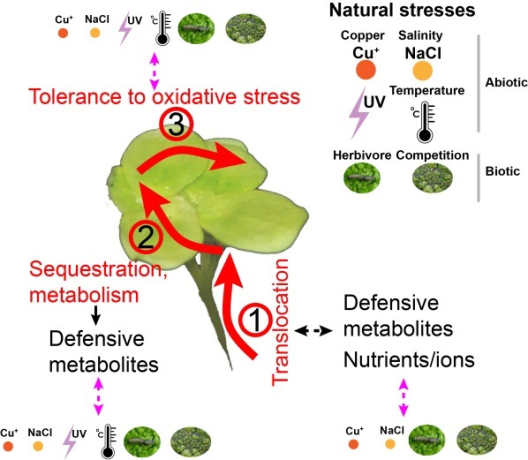
An emerging picture from recent studies suggests that adaptation to stress combinations, acting either simultaneously or sequentially, might differ from the adaptation to individual stress factors. However, the exact ecological conditions that shaped the adaptation in the past are difficult to know. Consequently, understanding the mechanisms and consequences of plant adaptation to environment changes represents a big challenge. To address this, investigating the process of adaptation-in-action, during which the process of adaptation can be monitored and environmental conditions are manipulated, is the best (if not the only) approach. In this project, using the giant duckweed – one of the fastest growing angiosperm plants, as a model system, we aim to investigate the process, mechanisms and consequences of adaptation to individual natural stresses, herbicides and their combinations. More specifically, we aim to achieve three main objectives: 1) characterize multi-generational genotype-phenotype-fitness adaptation maps in response to both individual and combinations of herbicide exposure and natural stresses (salt, copper and species competition); 2) examine to what extent adaptation to natural stress facilitates or constrains adaptive responses to herbicides exposure; 3) identify the contribution of phenotypic plasticity to adaptive evolution to individual and combinations of stresses. This integrative study will bridge a knowledge gap in understanding the molecular mechanisms, processes and consequences of adaptation to stress combination, as well as reveal the yet unknown molecular and evolutionary mechanisms of herbicide resistance and thus facilitate new strategies to improve weed management.
Key collaborators:
Prof. Alex Widmer, ETH Zurich, Switzerland
PD.Dr. Klaus Appenroth, Friedrich-Schiller-Universität Jena, Germany
Prof. Rensen Zeng, Fujian Agriculture and Forestry University, China