Team
- Johannes Holert
- Julia Wessel
- Yulduzkhon Abdullaeva
- N.N.
Topic
Synthetic compounds are constantly and increasingly entering our environment. These include plastic, pesticides, pharmaceuticals and all kinds of industrial chemicals. It is desirable to remove synthetic compounds from waste- and surface-waters via microbial degradation. Although this is achieved for many compounds microbial degradation of synthetic compounds is a permanent challenge because they are often very stable and can be toxic to microorganisms. For many synthetic compounds, metabolic pathways do not exist due because structurally similar natural compounds are lacking as templates.
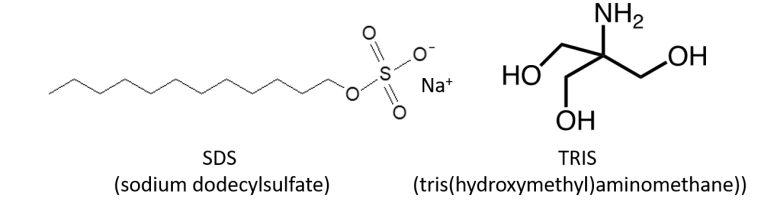
We are studying the degradation of synthetic chemicals with two model systems. The first is the degradation of the anionic surfactant SDS by Pseudomonas aeruginosa. As SDS is very toxic to bacteria, we found that the cells form cell aggregates and biofilms to protect themselves from these toxic effects. We also found that the degradation pathway involves a novel dehydrogenase for long-chain alcohols which is also required for the degradation of alkanes by this organism.
The second system is degradation of the synthetic buffer compound TRIS, which is abundantly used in biological laboratories and also as an ingredient of pharmaceuticals. It has been regarded as inert to biological degradation for a long time. By serendipity, we discovered TRIS-degrading bacteria in sewage treatment plants in the Münsterland region. The metabolic pathway consists of two gene cluster that catalyze the oxidation of TRIS to an amino acid and the further degradation of this intermediate, respectively. These two gene clusters are found in all TRIS-degrading strains so far and are mostly located on a large plasmid that can be transferred by horizontal gene transfer. Currently, the enzymes for the oxidation reactions as well as a transcriptional regulator are in the focus of our research.
In addition, we are studying microbial degradation processes in activated carbon filters used for advanced purification of wastewater.
© AG Philipp Funding
- BMWi (PaKuNaS)
- DFG (PH71/2-1)
Publications
Holert J, Borker A, Nübel LL, Daniel R, Poehlein A, Philipp B (2024) Bacteria use a catabolic patchwork pathway of apparently recent origin for degradation of the synthetic buffer compound TRIS
The ISME Journal, wrad023, https://doi.org/10.1093/ismejo/wrad023Panasia G, Drees SL, Fetzner S, Philipp B (2021) A complex of LaoA and LaoB acts as a Tat-dependent dehydrogenase for long-chain alcohols in Pseudomonas aeruginosa.
Appl. Environ. Microbiol. doi: 10.1128/AEM.00762-21Panasia G, Oetermann S, Steinbüchel A, Philipp B (2019) Sulfate ester detergent degradation in Pseudomonas aeruginosa is subject to both positive and negative regulation.
Applied and Environmental Microbiology 85(23): e01352-19. doi:10.1128/AEM.01352-19Panasia G, Philipp B (2018) Identification of LaoABCR as a novel system for the oxidation of long-chain alcohols derived from SDS- and alkane-degradation in Pseudomonas aeruginosa.
Applied and Environmental Microbiology 84: e00626-18. doi: 10.1128/AEM.00626-18Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B (2009) SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa.
Environmental Microbiology 11: 3073-3086. doi: 10.1111/j.1462-2920.2009.02012.xKlebensberger J, Lautenschlager K, Bressler D, Wingender J, Philipp B (2007) Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a pre-adaptive survival strategy.
Environmental Microbiology 9: 2247-2259. doi: 10.1111/j.1462-2920.2007.01339.xKlebensberger J, Rui O, Fritz E, Schink B, Philipp B (2006) Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate.
Archives of Microbiology 185: 417-427. doi: 10.1007/s00203-006-0111-y