Bacterial secondary metabolism:
enzyme-catalyzed reaction cascades towards bioactive natural productsMicroorganisms produce a wealth of bioactive compounds, many of which have been and continue to be crucial for medical, environmental or technological applications. This is reflected by the simple fact that, despite major efforts in pharmaceutical chemistry, the majority of registered drugs still originates from natural sources. Bacterial secondary metabolism also involves novel enzymes with unique properties, so that biosynthetic pathways can reveal exciting, uncharted reactions.
However, a multitude of biosynthetic gene clusters cannot be associated with particular products, probably due to lacking gene expression, precursor supply, instability of the respective pathway products, or analytical challenges.
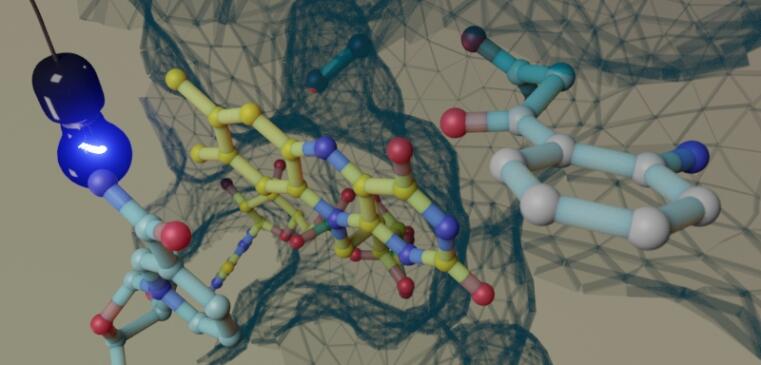
Working with the opportunistic pathogen Pseudomonas aeruginosa and other proteobacteria, we aim at the analysis of uncharacterized biosynthetic pathways in these bacteria, mainly using an enzyme-centered approach. Such a bottom-up strategy can provide useful hints on the identities of yet unknown metabolites or metabolite substructures. Reconstituting and assembling key reactions and reaction cascades occurring within these pathways discloses the fascinating mechanisms and properties of the enzymes involved. It can also provide useful details for the biotechnological, synthetic or semisynthetic production of metabolites for further applications.
Publications