Biochemical characterization of poly(cis-1,4-isoprene) degradation
 First studies on microbial degradation of natural rubber were already done about 100 years ago. However, until now only little is known about the molecular fundamentals and the biochemistry of poly(cis-1,4-isoprene) degradation. Poly(cis-1,4-isoprene) is the main constituent (> 90% of dry weight) of natural rubber (NR), a biopolymer synthesized by many plant species and some fungi in varying degrees of quality and quantity. For technical applications NR is obtained from the rubber tree Hevea brasiliensis (a). Alternatively, poly(cis-1,4-isoprene) (IR) is synthesized chemically from isoprene.
First studies on microbial degradation of natural rubber were already done about 100 years ago. However, until now only little is known about the molecular fundamentals and the biochemistry of poly(cis-1,4-isoprene) degradation. Poly(cis-1,4-isoprene) is the main constituent (> 90% of dry weight) of natural rubber (NR), a biopolymer synthesized by many plant species and some fungi in varying degrees of quality and quantity. For technical applications NR is obtained from the rubber tree Hevea brasiliensis (a). Alternatively, poly(cis-1,4-isoprene) (IR) is synthesized chemically from isoprene.
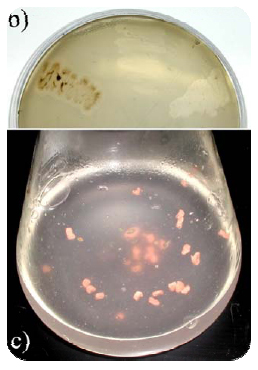 Two different groups of rubber degrading bacteria can be distinguished according to their decomposition strategies. Bacteria of the first group belong to the CMN group (Corynebacterium, Mycobacterium, Nocardia) and need direct physical contact to the rubber material showing adhesive growth. Several strains belonging to the genus Gordonia and Nocardia were identified as rubber-degrading bacteria. These strains show good rubber degradation activities in submers culture but do not grow and do not form clear zones on latex overlay agar plates containing rubber as sole carbon source (c). Members of the second group show weaker growth in submers culture, but grow and form clear zones on NR latex-overlay agar plates. These bacteria belong to streptomycetes and related genera. Streptomyces sp. K30 is one example (b). Only little is known about the biochemical mechanisms of rubber disintegration. An oxidative cleavage of the double bond in the polymer backbone for initiation of rubber degradation has been suggested by Tsuchii and Takeda, and diketone derivatives of oligo(cis-1,4-isoprene) were identified as metabolites of rubber degradation. Recently, two different enzymes capable of poly(cis-1,4-isoprene) cleavage were described: RoxA was isolated from Xanthomonas sp., and Lcp was identified in Streptomyces sp. K30 in our workinggroup. Recently, also enzyme mediator systems for in vitro oxidative degradation of polyisoprene were described, which are based on free radical chain reactions of lipids using different known enzymes like manganese peroxidase, laccase, horseradish peroxidase and lipoxigenase/linoleic acid. For the first time a gene was identified which is responsible for clear zone formation on latex and which is essential for the process of rubber degradation during clear zone formation by actinomycetes.
Two different groups of rubber degrading bacteria can be distinguished according to their decomposition strategies. Bacteria of the first group belong to the CMN group (Corynebacterium, Mycobacterium, Nocardia) and need direct physical contact to the rubber material showing adhesive growth. Several strains belonging to the genus Gordonia and Nocardia were identified as rubber-degrading bacteria. These strains show good rubber degradation activities in submers culture but do not grow and do not form clear zones on latex overlay agar plates containing rubber as sole carbon source (c). Members of the second group show weaker growth in submers culture, but grow and form clear zones on NR latex-overlay agar plates. These bacteria belong to streptomycetes and related genera. Streptomyces sp. K30 is one example (b). Only little is known about the biochemical mechanisms of rubber disintegration. An oxidative cleavage of the double bond in the polymer backbone for initiation of rubber degradation has been suggested by Tsuchii and Takeda, and diketone derivatives of oligo(cis-1,4-isoprene) were identified as metabolites of rubber degradation. Recently, two different enzymes capable of poly(cis-1,4-isoprene) cleavage were described: RoxA was isolated from Xanthomonas sp., and Lcp was identified in Streptomyces sp. K30 in our workinggroup. Recently, also enzyme mediator systems for in vitro oxidative degradation of polyisoprene were described, which are based on free radical chain reactions of lipids using different known enzymes like manganese peroxidase, laccase, horseradish peroxidase and lipoxigenase/linoleic acid. For the first time a gene was identified which is responsible for clear zone formation on latex and which is essential for the process of rubber degradation during clear zone formation by actinomycetes.
This project will address the functional and enzymatic characterization of Lcp homologous proteins identified in Streptomyces sp., Gordonia sp. and Nocardia sp. in respect to cleavage of poly(cis-1,4-isoprene). Furthermore, enzymatic steps catalyzed by OxiBA following poly(cis-1,4-isoprene) cleavage by Lcp will be investigated. Different various of genetic methods are used, e.g. DNA transfer techniques for heterologous expression, purification of the different enzymes, different protein techniques, enzyme assays.