Our group focuses on different compartmentalized (iso)enzymes in higher plants that either operate at the junction of primary and secondary metabolism or are involved in the post-translational modification of secreted glycoproteins.
Aims
The goal of our work is to understand the need for sub-cellular separation of comparable or successive enzymatic sequences that occurred gradually during evolution. We are especially interested in how metabolic flux is channelled by certain modifications or regulation levels. We thus aim at contributing to understand the extraordinary PLASTICITY of higher plant metabolism that reflects adaptation to an obligate sessile life style.
Indirect evidences
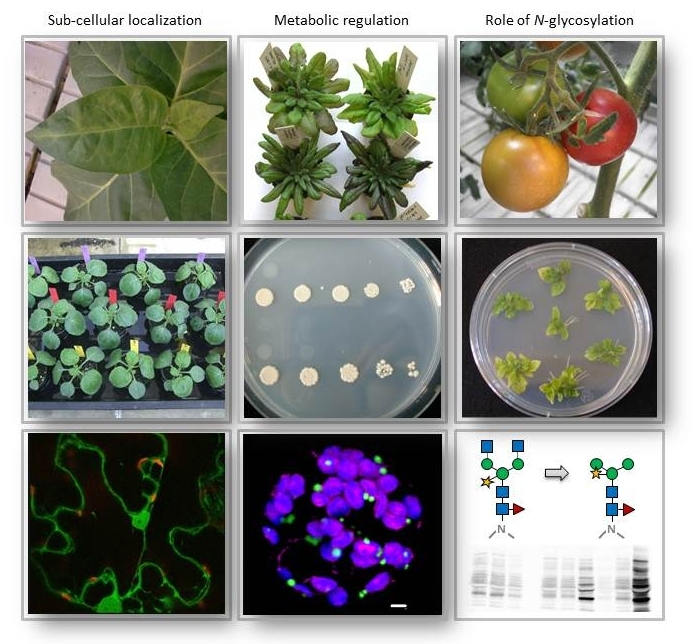
By measuring enzymatic activities or with the help of molecular probes we investigate predominately Arabidopsis and representatives of the Solanaceae (tobacco and tomato). Besides specific DNA- or RNA probes or isoform-specific antibodies, we more and more came to rely on life-cell imaging (confocal laser-scanning microscopy, CLSM). To be able to use this technique, we first clone genomic or cDNA fragments (the latter by RT-PCR from total RNA) and clone them in basic vectors prior to create different expression constructs (for E. coli or plant cells). Functional analyses encompass site-directed mutagenesis to specifically introduce base-changes resulting in single amino acid changes. The synthesis of recombinant proteins is conducted in special E. coli strains to enable either biochemical characterization in enzyme-deficient strains or to produce antisera in rabbits (after affinity-purification). The latter are usually purified to enhance specificity (e.g. depleted against other isoforms) and to avoid immunological cross reaction with related proteins.
Techniques
Targeting studies of isoform members is routinely done by reporter fusions that allow for sub-cellular localization, either due to a colour reaction or to emission of specific light/fluorescent signals. We also study dynamics of protein accumulation versus protein degradation (turnover) in leaf tissue upon Agrobacterium infiltration or in directly transfected protoplasts using suitable detection methods.
Arabidopsis KO mutants are used for most loss-of-function analyses. Stable knock-down in other plants is achieved by RNAi-mediated gene suppression after generating stably transformed lines (regeneration of sterile tissue explants = T0, up to T5). Experimental reduction or overexpression of single isoenzymes in whole plants is indispensable for elucidating their specific role and function in metabolism. If required, different lines are crossed to achieve a combination of single effects.
Protein-protein interactions are analysed with help of the ”yeast 2-hybrid” system or directly in plant cells (by BiFC or specific pull-down analyses). Evolutionary aspects (bioinformatics comparisons) complete our analyses.
