Chiral molecules on surfaces
Matthias Kettner, Daniel Nürenberg, Melanie Kämmerer
Chiral molecules are defined as molecules which lack mirror image symmetry. They have two types of enantiomers that can be described as left-handed and right-handed species. One example is the helical structure of double stranded (ds) DNA. - Enantioselectivity describes the effect that one enantiomer of a chiral product is preferentially produced in a chemical reaction. In living species Nature selects one type of enantiomer: left-handed amino acids, right handed sugars and right handed DNA. Within this project the interaction of chiral molecules and spin polarized electrons is investigated.
Thin films of chiral molecules are prepared in collaboration with the Weizmann Institute of Science in Israel via self-assembly: Double stranded DNA with a thiolated 3’ end as well as thiolated helical poly-amino acids is deposited on a gold surface. The sulfur binds to the gold surface, and as long as the length of the molecules is shorter than the persistent length one gets a layer of molecules standing like rigid rods on the surface. [1,2]
Other experiments are performed with purple membrane containing bacteriorhodopsin. This protein consists mainly of seven α-helices standing upright within the membrane. Molecules are physisorbed on metal surfaces. [3]
The spin polarization of photoelectrons emitted from the gold substrate and transmitted through the layer of chiral molecules is measured. For spin polarization measurements a conventional small-sized Mott polarimeter is applied. To ensure that the photoelectrons originate from the substrate, photon energy is chosen that is not sufficient to ionize the adsorbed molecules. Spin polarization is measured for different lengths of molecules adsorbed on different substrates, mainly a gold single crystal, poly-crystalline gold and aluminum samples to determine the influence of the substrates spin-orbit coupling. Photoelectrons are excited by light of different polarizations, either linearly or circularly polarized light.
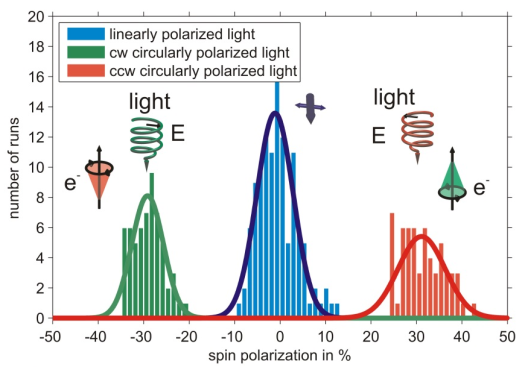
Using a bare gold substrate spin polarized photoelectrons can be excited by circularly polarized light [4], a measurement is shown in figure 1. The sign of the spin polarization can be reversed by changing between the two helicities of the photoexciting light.
It is well known that photoelectrons excited from a gold (111) surface by circularly polarized light are spin-polarized due to spin selective transitions contributing to the spectrum [5]. In this context the measurement serves as a proof of principle of the experimental apparatus.
ToF measurements
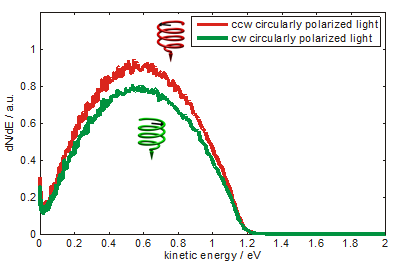
Photoelectron yield - excited from a coated gold surface, as indicated on the left - depends upon the polarization of photoemitting light as shown in Fig.1. Spectra are measured using an electron time-of-flight detector and pulsed laser radiation of 5.8 eV photon energy. The difference of the photoelectron yield is constant over the measured range of photoelectron kinetic energy. About 5% asymmetry is calculated from the spectrum (fig. 2). The preference of electrons photoemitted by counterclockwise circularly polarized light is interpreted as a preferential transmission of electrons by the adsorbed DNA molecules. As indicated in the sketch the molecules are standing like rods on the surface acting as a spin filter.
Spin-resolved measurements
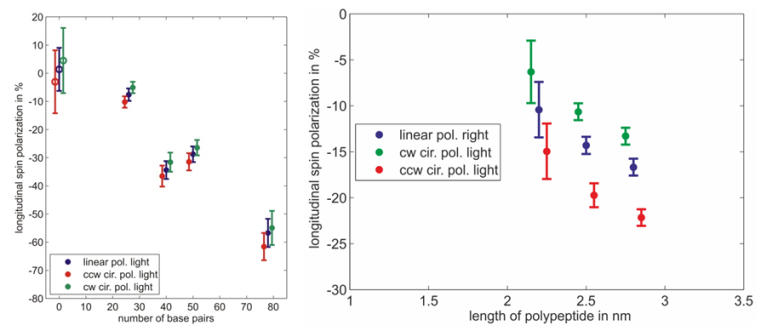
Spin polarimetry measurements show that helical molecules adsorbed on gold surfaces indeed act as an efficient spin filter for electrons. Furthermore the resulting spin polarization depends on the length of the adsorbed molecules and the quality of the monolayer. The measured spin polarization increases with the length of the molecules (fig. 3). Spin polarization measured on dsDNA functionalized gold surfaces is larger than in case of polypeptides whereas the molecules are small compared to dsDNA.
Moreover a systematic study reveals that the spin polarization decreases if the monolayer is not well organized. This is shown by adsorbing single stranded DNA molecules which form a rather floppy than rigid layer. Electrons transmitted through the layer of single stranded DNA molecules are not significantly spin polarized. In another experiment monolayers of double stranded DNA have been damaged intentionally with intense UV light. In this case, the spin polarization decreases with increasing exposure time to zero [7].
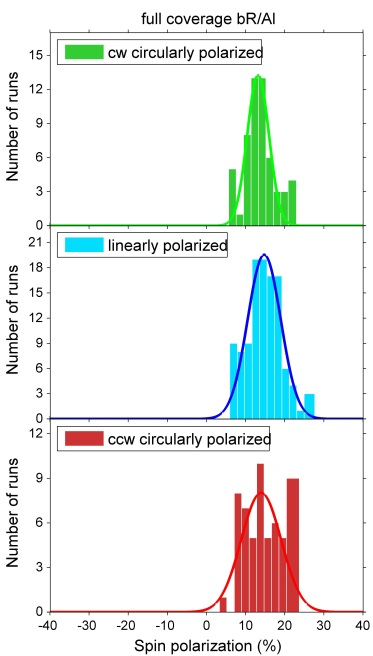
Investigations of bacteriorhodopsin (bR) physisorbed on metal surfaces result in absolute electron spin polarization with similar magnitude as in the case of polypeptides (fig. 4). Due to the physisorption one can exclude influences of bonds between molecules and surfaces. By changing the substrate it is furthermore possible to determine the influence of the spin orbit coupling.
Mott polarimeter
A Mott polarimeter is a device to measure electron spin polarization utilizing spin orbit interaction. In this setup photoelectrons are accelerated by 50kV onto a thin gold foil. Fast electrons are scattered within the Coulomb field of the nuclei of the target. Within the rest frame of the electron the scattering nucleus can be seen as a moving accelerated charge resulting in a magnetic field which interacts with the electrons’ spin. Therefore, the interaction potential depends on the orientation of the electrons’ spin and orbit of motion. A symmetric setup of two detector counting scattered electrons – under certain angles [6] – will measure an asymmetry
where Nl and Nr denote the number of scattered electrons to the left and right detector, respectively. This asymmetry is proportional to the spin polarization of incident electrons. The spin-orbit interaction is quantified by the analyzing power Seff, also known as the Sherman function. The transversal polarization P is then calculated by
In this experiment a conventional electron polarimeter of a compact size operating at 50 kV is used, the Sherman function Seff has been calibrated by determining the left-right asymmetry of an electron beam with a well-known spin polarization at the group of G. F. Hanne.
Literature
[1] S. G. Ray, S. S. Daube, G. Leitus, Z. Vager, and R. Naaman PRL 96, 036101, 2006
[2] M. Kettner B. Göhler, H. Zacharias, D. Mishra, V. Kiran, R. Naaman, C. Fontanesi, D. H. Waldeck, S. Sęk, J. Pawłowski, J. Juhaniewicz, JPPC, ASAP
[3] D. Mishra, T. Z., R. Naamana, M. Kettner, B. Göhler, H. Zacharias, N. Friedman, M. Sheves, and C. Fontanesi, PNAS 110, 37, 2013
[4] F. Meier and D. Pescia, PRL 47, 374, 1981
[5] G. Borstel and M. Wöhlecke, PRB 26, 1148, 1982
[6] Polarized Electrons, J. Kessler, Springer
[7] B. Göhler, V. Hamelbeck, T. Z. Markus, M. Kettner, G. F. Hanne, Z. Vager, R. Naaman, and H. Zacharias, Science 331, 894, 2011


